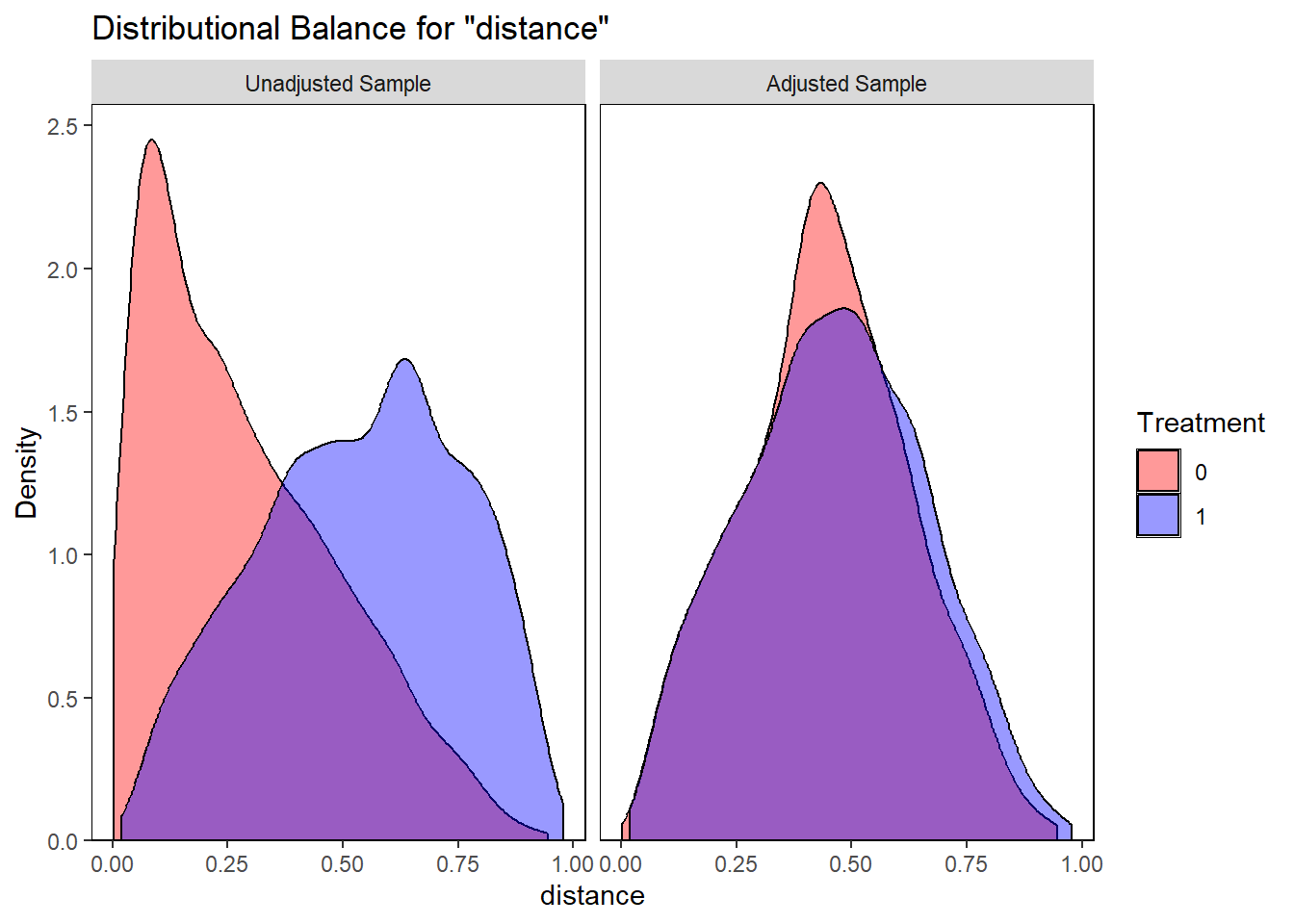
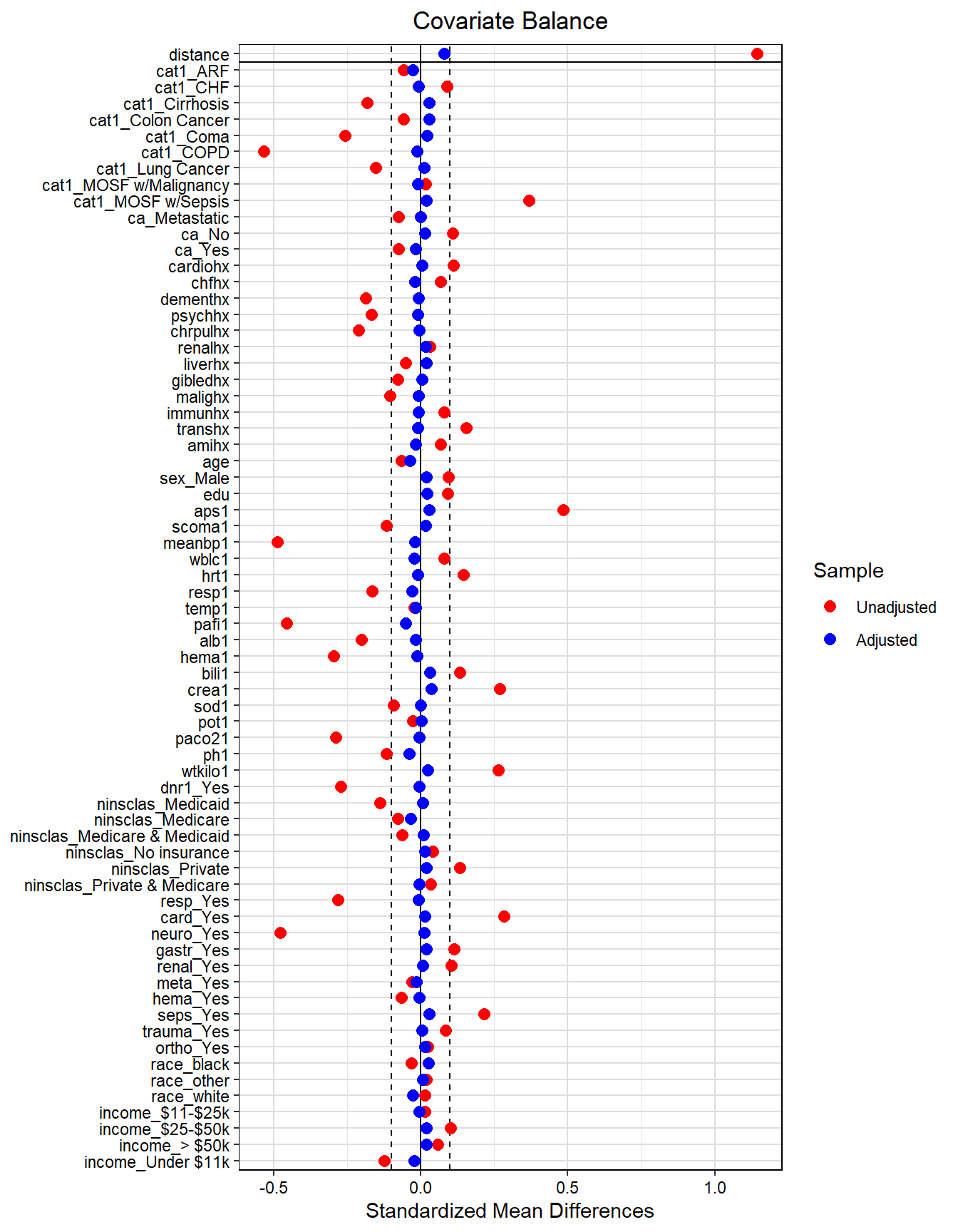
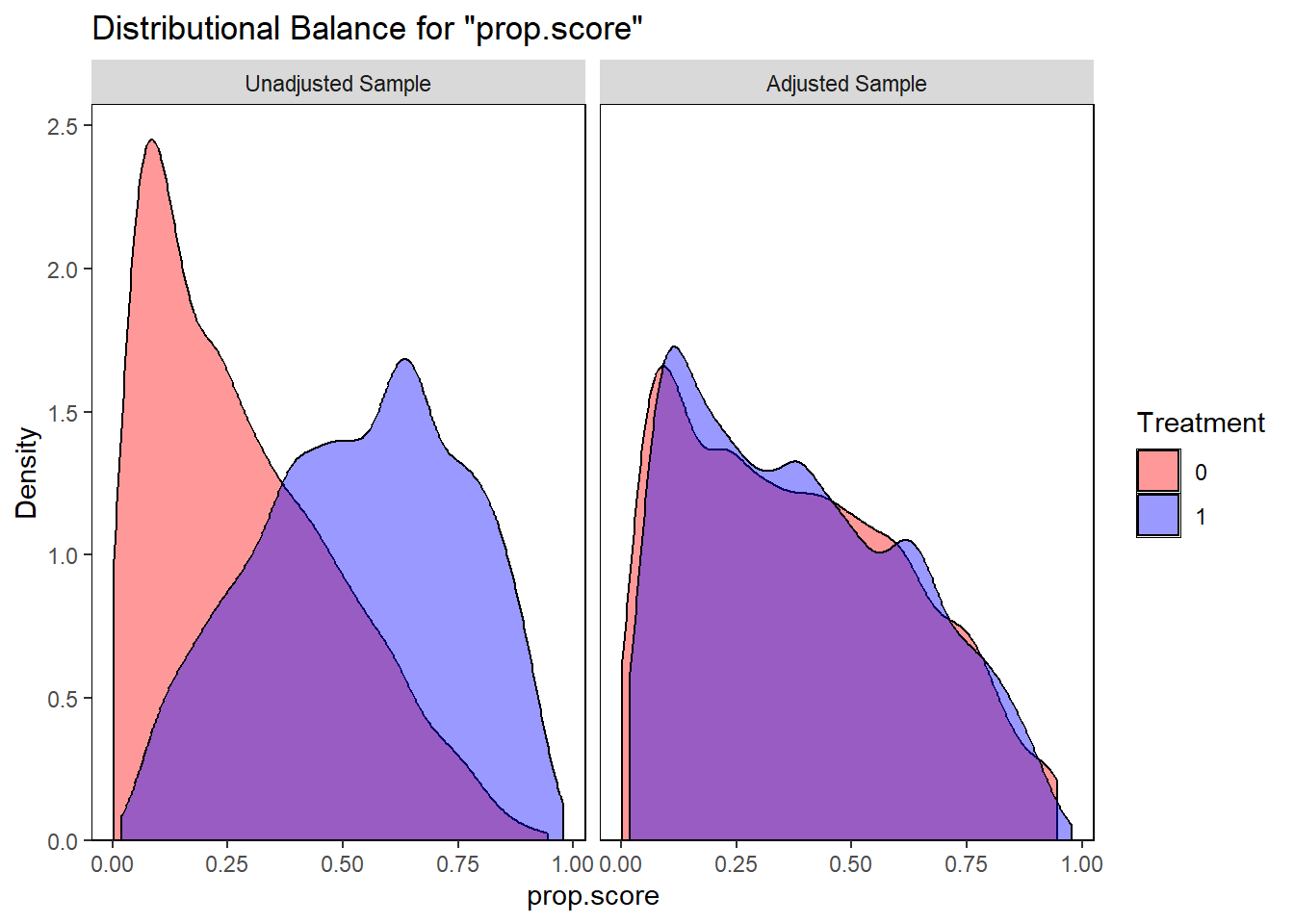
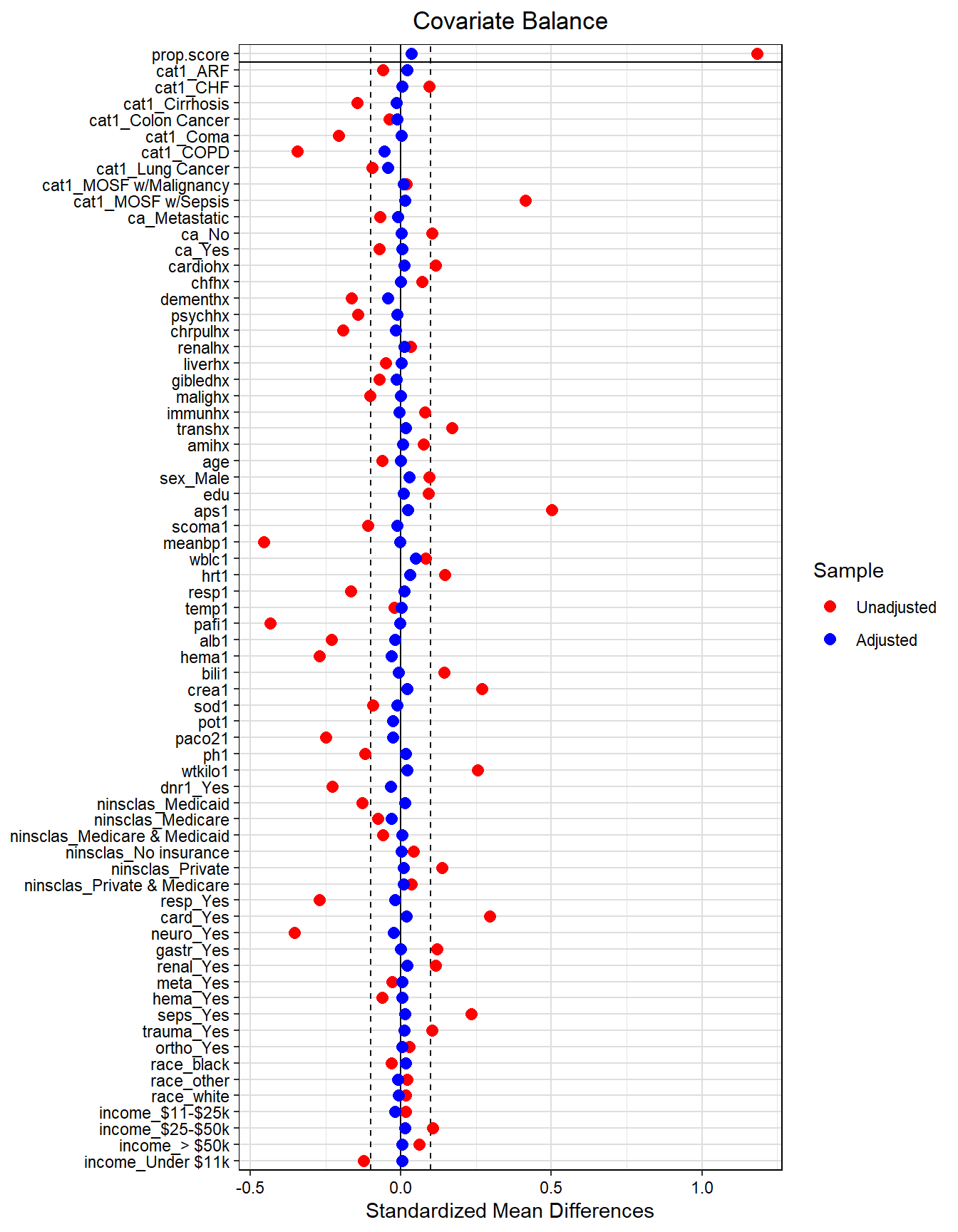
require(tidyverse)
library(tableone)
data2<-readRDS("data/data2")
covariates <- select(data2, -c(id, A, Y))
baselines <- colnames(covariates)
baselines [1] "cat1" "ca" "cardiohx" "chfhx" "dementhx" "psychhx"
[7] "chrpulhx" "renalhx" "liverhx" "gibledhx" "malighx" "immunhx"
[13] "transhx" "amihx" "age" "sex" "edu" "aps1"
[19] "scoma1" "meanbp1" "wblc1" "hrt1" "resp1" "temp1"
[25] "pafi1" "alb1" "hema1" "bili1" "crea1" "sod1"
[31] "pot1" "paco21" "ph1" "wtkilo1" "dnr1" "ninsclas"
[37] "resp" "card" "neuro" "gastr" "renal" "meta"
[43] "hema" "seps" "trauma" "ortho" "race" "income" tab0 <- CreateTableOne(vars = baselines,
data = data2,
strata = "A",
test = FALSE, #mute P-value calculation;
smd = TRUE,
addOverall = TRUE)
print(tab0, smd = TRUE, showAllLevels = FALSE) Stratified by A
Overall 0 1 SMD
n 5735 3551 2184
cat1 (%) 0.583
ARF 2490 (43.4) 1581 (44.5) 909 (41.6)
CHF 456 ( 8.0) 247 ( 7.0) 209 ( 9.6)
Cirrhosis 224 ( 3.9) 175 ( 4.9) 49 ( 2.2)
Colon Cancer 7 ( 0.1) 6 ( 0.2) 1 ( 0.0)
Coma 436 ( 7.6) 341 ( 9.6) 95 ( 4.3)
COPD 457 ( 8.0) 399 (11.2) 58 ( 2.7)
Lung Cancer 39 ( 0.7) 34 ( 1.0) 5 ( 0.2)
MOSF w/Malignancy 399 ( 7.0) 241 ( 6.8) 158 ( 7.2)
MOSF w/Sepsis 1227 (21.4) 527 (14.8) 700 (32.1)
ca (%) 0.107
Metastatic 384 ( 6.7) 261 ( 7.4) 123 ( 5.6)
No 4379 (76.4) 2652 (74.7) 1727 (79.1)
Yes 972 (16.9) 638 (18.0) 334 (15.3)
cardiohx (mean (SD)) 0.18 (0.38) 0.16 (0.37) 0.20 (0.40) 0.116
chfhx (mean (SD)) 0.18 (0.38) 0.17 (0.37) 0.19 (0.40) 0.069
dementhx (mean (SD)) 0.10 (0.30) 0.12 (0.32) 0.07 (0.25) 0.163
psychhx (mean (SD)) 0.07 (0.25) 0.08 (0.27) 0.05 (0.21) 0.143
chrpulhx (mean (SD)) 0.19 (0.39) 0.22 (0.41) 0.14 (0.35) 0.192
renalhx (mean (SD)) 0.04 (0.21) 0.04 (0.20) 0.05 (0.21) 0.032
liverhx (mean (SD)) 0.07 (0.26) 0.07 (0.26) 0.06 (0.24) 0.049
gibledhx (mean (SD)) 0.03 (0.18) 0.04 (0.19) 0.02 (0.16) 0.070
malighx (mean (SD)) 0.23 (0.42) 0.25 (0.43) 0.20 (0.40) 0.101
immunhx (mean (SD)) 0.27 (0.44) 0.26 (0.44) 0.29 (0.45) 0.080
transhx (mean (SD)) 0.12 (0.32) 0.09 (0.29) 0.15 (0.36) 0.170
amihx (mean (SD)) 0.03 (0.18) 0.03 (0.17) 0.04 (0.20) 0.074
age (mean (SD)) 61.38 (16.68) 61.76 (17.29) 60.75 (15.63) 0.061
sex = Male (%) 3192 (55.7) 1914 (53.9) 1278 (58.5) 0.093
edu (mean (SD)) 11.68 (3.15) 11.57 (3.13) 11.86 (3.16) 0.091
aps1 (mean (SD)) 54.67 (19.96) 50.93 (18.81) 60.74 (20.27) 0.501
scoma1 (mean (SD)) 21.00 (30.27) 22.25 (31.37) 18.97 (28.26) 0.110
meanbp1 (mean (SD)) 78.52 (38.05) 84.87 (38.87) 68.20 (34.24) 0.455
wblc1 (mean (SD)) 15.65 (11.87) 15.26 (11.41) 16.27 (12.55) 0.084
hrt1 (mean (SD)) 115.18 (41.24) 112.87 (40.94) 118.93 (41.47) 0.147
resp1 (mean (SD)) 28.09 (14.08) 28.98 (13.95) 26.65 (14.17) 0.165
temp1 (mean (SD)) 37.62 (1.77) 37.63 (1.74) 37.59 (1.83) 0.021
pafi1 (mean (SD)) 222.27 (114.95) 240.63 (116.66) 192.43 (105.54) 0.433
alb1 (mean (SD)) 3.09 (0.78) 3.16 (0.67) 2.98 (0.93) 0.230
hema1 (mean (SD)) 31.87 (8.36) 32.70 (8.79) 30.51 (7.42) 0.269
bili1 (mean (SD)) 2.27 (4.80) 2.00 (4.43) 2.71 (5.33) 0.145
crea1 (mean (SD)) 2.13 (2.05) 1.92 (2.03) 2.47 (2.05) 0.270
sod1 (mean (SD)) 136.77 (7.66) 137.04 (7.68) 136.33 (7.60) 0.092
pot1 (mean (SD)) 4.07 (1.03) 4.08 (1.04) 4.05 (1.01) 0.027
paco21 (mean (SD)) 38.75 (13.18) 39.95 (14.24) 36.79 (10.97) 0.249
ph1 (mean (SD)) 7.39 (0.11) 7.39 (0.11) 7.38 (0.11) 0.120
wtkilo1 (mean (SD)) 67.83 (29.06) 65.04 (29.50) 72.36 (27.73) 0.256
dnr1 = Yes (%) 654 (11.4) 499 (14.1) 155 ( 7.1) 0.228
ninsclas (%) 0.194
Medicaid 647 (11.3) 454 (12.8) 193 ( 8.8)
Medicare 1458 (25.4) 947 (26.7) 511 (23.4)
Medicare & Medicaid 374 ( 6.5) 251 ( 7.1) 123 ( 5.6)
No insurance 322 ( 5.6) 186 ( 5.2) 136 ( 6.2)
Private 1698 (29.6) 967 (27.2) 731 (33.5)
Private & Medicare 1236 (21.6) 746 (21.0) 490 (22.4)
resp = Yes (%) 2113 (36.8) 1481 (41.7) 632 (28.9) 0.270
card = Yes (%) 1931 (33.7) 1007 (28.4) 924 (42.3) 0.295
neuro = Yes (%) 693 (12.1) 575 (16.2) 118 ( 5.4) 0.353
gastr = Yes (%) 942 (16.4) 522 (14.7) 420 (19.2) 0.121
renal = Yes (%) 295 ( 5.1) 147 ( 4.1) 148 ( 6.8) 0.116
meta = Yes (%) 265 ( 4.6) 172 ( 4.8) 93 ( 4.3) 0.028
hema = Yes (%) 354 ( 6.2) 239 ( 6.7) 115 ( 5.3) 0.062
seps = Yes (%) 1031 (18.0) 515 (14.5) 516 (23.6) 0.234
trauma = Yes (%) 52 ( 0.9) 18 ( 0.5) 34 ( 1.6) 0.104
ortho = Yes (%) 7 ( 0.1) 3 ( 0.1) 4 ( 0.2) 0.027
race (%) 0.036
black 920 (16.0) 585 (16.5) 335 (15.3)
other 355 ( 6.2) 213 ( 6.0) 142 ( 6.5)
white 4460 (77.8) 2753 (77.5) 1707 (78.2)
income (%) 0.142
$11-$25k 1165 (20.3) 713 (20.1) 452 (20.7)
$25-$50k 893 (15.6) 500 (14.1) 393 (18.0)
> $50k 451 ( 7.9) 257 ( 7.2) 194 ( 8.9)
Under $11k 3226 (56.3) 2081 (58.6) 1145 (52.4)