2. Using machine learning techniques for causal analysis
Outlines
- Propensity score methods machine learning techniques
- gradient boosting machines
- super learner
- Bayesian additive regression trees
Proposensity score analysis using machine learning technqiues
2.1 Super (Machine) Learning
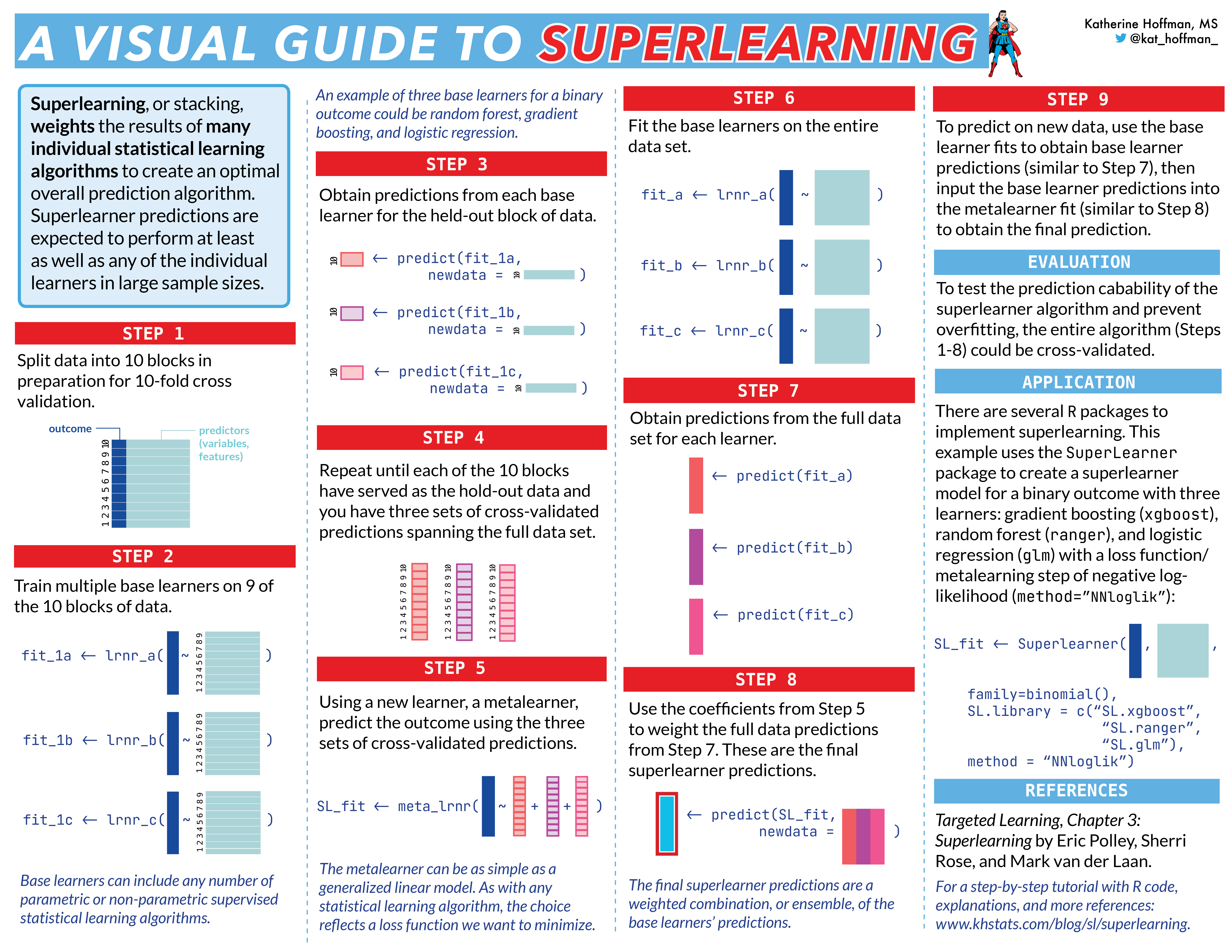
Super learning can be used to obtain robust estimator. In a nut-shell it uses loss-function based ML tool and cross-validation to obtain the best prediction of model parameter of interest, based on a weighted average of a library of machine learning algorithms.
Guide to SuperLearner by Chris Kennedy at https://cran.r-project.org/web/packages/SuperLearner/vignettes/Guide-to-SuperLearner.html
New visual guide created by Katherine Hoffman
- List of machine learning algorithms under SuperLearner R package
library(SuperLearner)
listWrappers() [1] "SL.bartMachine" "SL.bayesglm" "SL.biglasso"
[4] "SL.caret" "SL.caret.rpart" "SL.cforest"
[7] "SL.earth" "SL.gam" "SL.gbm"
[10] "SL.glm" "SL.glm.interaction" "SL.glmnet"
[13] "SL.ipredbagg" "SL.kernelKnn" "SL.knn"
[16] "SL.ksvm" "SL.lda" "SL.leekasso"
[19] "SL.lm" "SL.loess" "SL.logreg"
[22] "SL.mean" "SL.nnet" "SL.nnls"
[25] "SL.polymars" "SL.qda" "SL.randomForest"
[28] "SL.ranger" "SL.ridge" "SL.rpart"
[31] "SL.rpartPrune" "SL.speedglm" "SL.speedlm"
[34] "SL.step" "SL.step.forward" "SL.step.interaction"
[37] "SL.stepAIC" "SL.svm" "SL.template"
[40] "SL.xgboost" [1] "All"
[1] "screen.corP" "screen.corRank" "screen.glmnet"
[4] "screen.randomForest" "screen.SIS" "screen.template"
[7] "screen.ttest" "write.screen.template"2.2 Using machine learning methods with PSA
- We can use ML to model our propensity score model
- The use of machine learning methods is more flexible than parametric methods (i.e., logistic regression)
- Not without a cost, usually the more flexible the methods are the more one is at risk of overfitting; Too much noise considered in the modelling often results in poor coverage probability.
- There is no one approach that out performs others, thus which approach to use should be evaluated case by case.
- ML is generally suggested for large enough cohort and for modelling large set of covariates.
- It’s always suggested to include results from conventional logistic regression approach as a sensitivity analysis in comparison of ML approaches.
- Many approaches are included in the WeightIt package, https://ngreifer.github.io/WeightIt/reference/method_super.html
- “gbm”, Propensity score weighting using generalized boosted modeling (also known as gradient boosting machines)
- “super”, Propensity score weighting using SuperLearner
- “bart”, Propensity score weighting using Bayesian additive regression trees (BART)
- Bayesian Additive Regression Trees (BART) is a sum-of-trees model for approximating an unknown function. To avoid overfitting (of decision tree), BART uses a regularization prior that forces each tree to be able to explain only a limited subset of the relationships between the covariates and the predictor variable.
Setting up data and PS model formula
require(tidyverse)
require(WeightIt)
data2<-readRDS("data/data2")
covariates <- select(data2, -c(id, A, Y))
baselines <- colnames(covariates)
ps.formula <- as.formula(paste("A~",
paste(baselines, collapse = "+")))2.2.1 PS model with gradient boosting
- computationally more demanding and it might take several minutes to run.
IPTW_gbm <- weightit(ps.formula,
data = data2,
method = "gbm",
stabilize = TRUE)
# saving the model output as a R object to avoid rerunning the same model;
saveRDS(IPTW_gbm, file = "data/IPTW_gbm")# reading saved model output;
require(sjPlot)
IPTW_gbm <- readRDS(file = "data/IPTW_gbm")
summary(IPTW_gbm) Summary of weights
- Weight ranges:
Min Max
treated 0.3874 |---------------------------| 10.9875
control 0.6227 |------------| 5.6764
- Units with the 5 most extreme weights by group:
5525 3148 4890 1825 5131
treated 4.0556 4.6873 5.1931 5.3152 10.9875
497 2046 3830 1602 1000
control 4.6689 4.8977 4.929 4.9313 5.6764
- Weight statistics:
Coef of Var MAD Entropy # Zeros
treated 0.663 0.397 0.143 0
control 0.426 0.262 0.067 0
- Effective Sample Sizes:
Control Treated
Unweighted 3551. 2184.
Weighted 3006.24 1517.47fit2_gbm <- glm_weightit(Y ~ A,
family = "binomial",
weightit = IPTW_gbm,
data = data2)
tab_model(fit2_gbm)| Y | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.46 | 0.42 – 0.49 | <0.001 |
| A | 1.27 | 1.11 – 1.44 | <0.001 |
| Observations | 5735 | ||
| R2 Tjur | 0.004 | ||
2.2.2 PS model with Super Learner
IPTW_SL <- weightit(ps.formula,
data = data2,
method = "super",
SL.library=c("SL.randomForest", "SL.glmnet", "SL.nnet"),
stabilize = TRUE)
# saving the model output as a R object to avoid rerunning the same model;
saveRDS(IPTW_SL, file = "data/IPTW_SL")# reading saved model output;
IPTW_SL <- readRDS(file = "data/IPTW_SL")
summary(IPTW_SL) Summary of weights
- Weight ranges:
Min Max
treated 0.4008 |-----------------| 1.0346
control 0.6254 |--------------------| 1.4140
- Units with the 5 most extreme weights by group:
742 5131 3508 1825 4890
treated 0.9223 0.9489 0.968 0.9808 1.0346
3391 3216 2174 2986 505
control 1.3373 1.3461 1.3533 1.3627 1.414
- Weight statistics:
Coef of Var MAD Entropy # Zeros
treated 0.184 0.147 0.016 0
control 0.159 0.125 0.012 0
- Effective Sample Sizes:
Control Treated
Unweighted 3551. 2184.
Weighted 3463.76 2112.6fit2_SL <- glm_weightit(Y ~ A,
family = "binomial",
weightit = IPTW_SL,
data = data2)
tab_model(fit2_SL)| Y | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.45 | 0.42 – 0.48 | <0.001 |
| A | 1.35 | 1.21 – 1.51 | <0.001 |
| Observations | 5735 | ||
| R2 Tjur | 0.005 | ||
2.2.3 PS model with Bayesian additive regression trees
- A much faster algorithm comparing to gbm and SL.
IPTW_bart <- weightit(ps.formula,
data = data2,
method = "bart",
stabilize = TRUE)
# saving the model output as a R object to avoid rerunning the same model;
saveRDS(IPTW_bart, file = "data/IPTW_bart")# reading saved model output;
IPTW_bart <- readRDS(file = "data/IPTW_bart")
summary(IPTW_bart) Summary of weights
- Weight ranges:
Min Max
treated 0.3861 |---------------------------| 13.8560
control 0.6211 |------------------| 9.8333
- Units with the 5 most extreme weights by group:
3148 4119 1825 5131 4890
treated 8.7361 9.5095 11.8258 12.7766 13.856
84 1810 3839 1000 505
control 6.7109 6.7279 7.9066 7.9218 9.8333
- Weight statistics:
Coef of Var MAD Entropy # Zeros
treated 0.926 0.471 0.226 0
control 0.544 0.308 0.096 0
- Effective Sample Sizes:
Control Treated
Unweighted 3551. 2184.
Weighted 2740.56 1175.59fit2_bart <- glm_weightit(Y ~ A,
family = "binomial",
weightit = IPTW_bart,
data = data2)
tab_model(fit2_bart)| Y | |||
| Predictors | Odds Ratios | CI | p |
| (Intercept) | 0.45 | 0.42 – 0.49 | <0.001 |
| A | 1.30 | 1.13 – 1.50 | <0.001 |
| Observations | 5735 | ||
| R2 Tjur | 0.005 | ||
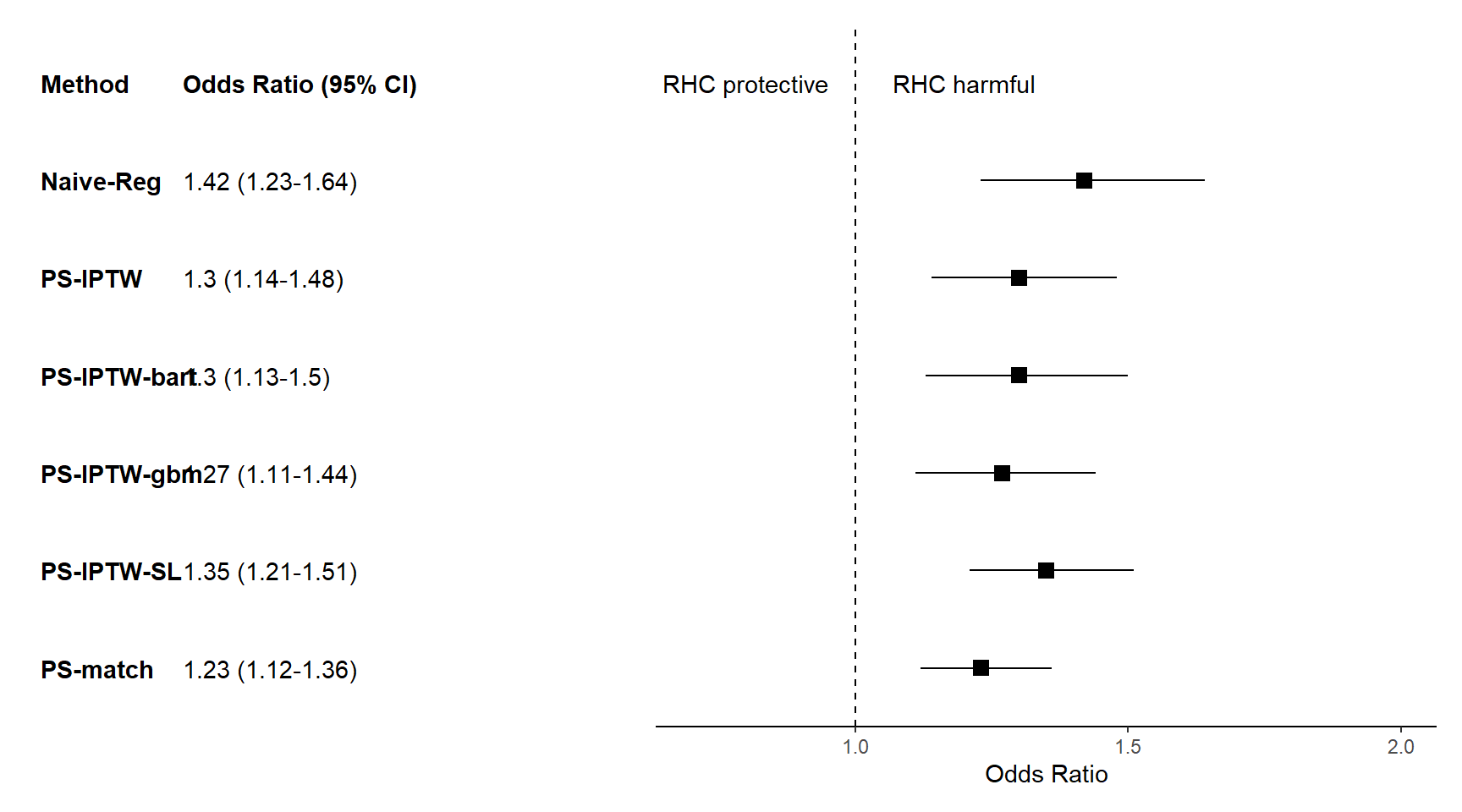
Comparing the three approaches given the rhc data, it appears that SuperLearner returns good stable weights (no visible extreme weights). This indicates a great fit of the PS model.
- Similar as before, we can check for PS distribution and balancing. Additionally, we can perform subgroup and sensitivity analysis as mentioned before.
Forest plot to display results from different approaches
- code modified from https://www.khstats.com/blog/forest-plots/ by Katherine Hoffman
plotdata <- data.frame(
method = c("Naive-Reg","PS-match", "PS-IPTW", "PS-IPTW-gbm", "PS-IPTW-SL", "PS-IPTW-bart"),
est.OR = c(1.42, 1.23, 1.30, 1.27, 1.35, 1.30),
conf.low = c(1.23, 1.12, 1.14, 1.11, 1.21, 1.13),
conf.high = c(1.64, 1.36, 1.48, 1.44, 1.51, 1.50))
p <-
plotdata |>
ggplot(aes(y = fct_rev(method))) +
theme_classic() +
geom_point(aes(x=est.OR), shape=15, size=3) +
geom_linerange(aes(xmin=conf.low, xmax=conf.high)) +
geom_vline(xintercept = 1, linetype="dashed") +
labs(x="Odds Ratio", y="") +
coord_cartesian(ylim=c(1,7), xlim=c(0.7, 2)) +
annotate("text", x = 0.8, y = 7, label = "RHC protective") +
annotate("text", x = 1.2, y = 7, label = "RHC harmful") +
theme(axis.line.y = element_blank(),
axis.ticks.y= element_blank(),
axis.text.y= element_blank(),
axis.title.y= element_blank())
plotdata_OR <- plotdata |>
# round estimates and 95% CIs to 2 decimal places for journal specifications
mutate(estimate_label = paste0(est.OR, " (", conf.low, "-", conf.high, ")")) |>
# add a row of data to be shown on the forest plot as column names;
bind_rows(
data.frame(
method = "Method",
estimate_label = "Odds Ratio (95% CI)"
)
) |>
mutate(method = fct_rev(fct_relevel(method, "Method")))
p_left <-
plotdata_OR |>
ggplot(aes(y = method)) +
geom_text(aes(x = 0, label = method), hjust = 0, fontface = "bold")+
geom_text(
aes(x = 1, label = estimate_label),
hjust = 0,
fontface = ifelse(plotdata_OR$estimate_label == "Odds Ratio (95% CI)", "bold", "plain")
)+
theme_void() +
coord_cartesian(xlim = c(0, 4))
library(patchwork)
layout <- c(
area(t = 0, l = 0, b = 14, r = 4), # left plot, starts at the top of the page (0) and goes 14 units down and 4 units to the right;
area(t = 1, l = 5, b = 14, r = 9) # middle plot starts a little lower (t=1) because there's no title. starts 1 unit right of the left plot (l=5, whereas left plot is r=4);
)
# final plot arrangement
p_left + p +plot_layout(design = layout)