library(tidyverse)
library(gtsummary)
library(DT)
options(scipen = 999)
indo <- read.csv("indo.csv", header = TRUE,fileEncoding="UTF-8-BOM")
# look at new data;
datatable(indo, rownames = FALSE) Heterogeneous Treatment Effects (HTE)
1. Hands-on excerise carrying out a simple risk-based HTE analysis
1.1 Data
Indomethacin for the Prevention of Post-ERCP Pancreatitis Dataset Description Document
This data set is provided by B. Joseph Elmunzer, the primary author of a study published on 2012 in the New England Journal of Medicine, volume 366, pages 1414-1422.
This RCT data contains baseline characteristics and outcomes for 602 participants at increased risk of post-ERCP pancreatitis at 4 centers.
We will be using a modified version of this data
1.2 Fitting a outcome logistic regression without treatment
- Calculate risk score and risk groups
model <- glm(outcome ~ ., data = indo[, c(-1, -2)], family = binomial)
summary(model)
Call:
glm(formula = outcome ~ ., family = binomial, data = indo[, c(-1,
-2)])
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -3.717127 1.014570 -3.664 0.000249 ***
age 0.002292 0.011126 0.206 0.836758
risk 0.998877 0.447973 2.230 0.025763 *
gender 0.104782 0.359077 0.292 0.770432
sod -0.622085 0.508628 -1.223 0.221305
pep 0.077874 0.526669 0.148 0.882452
recpanc -0.457586 0.358885 -1.275 0.202303
psphinc -0.330524 0.533223 -0.620 0.535349
precut -0.898827 0.672453 -1.337 0.181340
difcan -0.741625 0.513565 -1.444 0.148719
paninj -0.122011 0.446974 -0.273 0.784875
asa -0.025352 0.501523 -0.051 0.959684
sodsom -0.552573 0.357454 -1.546 0.122139
bsphinc 0.178201 0.288961 0.617 0.537435
bstent 0.015613 0.542894 0.029 0.977057
chole 0.932204 0.558188 1.670 0.094908 .
train 0.608712 0.265496 2.293 0.021863 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 467.73 on 600 degrees of freedom
Residual deviance: 437.13 on 584 degrees of freedom
AIC: 471.13
Number of Fisher Scoring iterations: 5# Calculate predicted risk score;
logOR <- model.matrix(model) %*% coef(model)
logOR_mean <- sum(coef(model)*model$means)
risk_score <- logOR - logOR_mean # centred PI;
indo$risk_score <- risk_score
# Rank patients by risk score and group into Quintile;
indo$riskgp <- cut(
indo$risk_score,
breaks = quantile(indo$risk_score, probs = seq(0, 1, by = 0.2)),
include.lowest = TRUE,
labels = 1:5
)1.3 Fitting a outcome logistic regression with treatment and risk group
trtmodel <- glm(outcome ~ rx*riskgp, data = indo, family = binomial)
summary(trtmodel)
Call:
glm(formula = outcome ~ rx * riskgp, family = binomial, data = indo)
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) -2.9957 0.5916 -5.064 0.000000411 ***
rx -1.0473 1.1693 -0.896 0.370413
riskgp2 0.9480 0.7008 1.353 0.176125
riskgp3 1.1350 0.7177 1.581 0.113779
riskgp4 1.7636 0.6650 2.652 0.008004 **
riskgp5 2.3026 0.6519 3.532 0.000412 ***
rx:riskgp2 0.6527 1.3342 0.489 0.624712
rx:riskgp3 0.3744 1.3222 0.283 0.777062
rx:riskgp4 0.1200 1.2827 0.094 0.925479
rx:riskgp5 0.2465 1.2464 0.198 0.843200
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
(Dispersion parameter for binomial family taken to be 1)
Null deviance: 467.73 on 600 degrees of freedom
Residual deviance: 427.03 on 591 degrees of freedom
AIC: 447.03
Number of Fisher Scoring iterations: 6# Calculate risk group-specific treatment effect on the OR scale;
# Extract coefficients and variance-covariance matrix
coefficients <- coef(trtmodel)
vcov_matrix <- vcov(trtmodel)
# Initialize a data frame to store results
results <- data.frame(
riskgp = levels(indo$riskgp),
logOR = NA,
SE = NA,
OR = NA,
lower_CI = NA,
upper_CI = NA
)
# Loop through each group
for (i in 1:length(levels(indo$riskgp))) {
grp <- levels(indo$riskgp)[i]
# Create a contrast vector
contrast <- rep(0, length(coefficients))
names(contrast) <- names(coefficients)
# Main effect of rx
contrast["rx"] <- 1
# Add interaction term if not the reference group
if (grp != levels(indo$riskgp)[1]) {
interaction_term <- paste0("rx:riskgp", grp)
contrast[interaction_term] <- 1
}
# Compute the log odds ratio
logOR <- sum(contrast * coefficients)
# Compute the standard error
SE <- sqrt(t(contrast) %*% vcov_matrix %*% contrast)
# Compute the odds ratio and confidence intervals
OR <- exp(logOR)
lower_CI <- exp(logOR - 1.96 * SE)
upper_CI <- exp(logOR + 1.96 * SE)
# Store the results
results$logOR[i] <- logOR
results$SE[i] <- SE
results$OR[i] <- OR
results$lower_CI[i] <- lower_CI
results$upper_CI[i] <- upper_CI
}
# Plot the group-specific rx effects
ggplot(results, aes(x = OR, y = riskgp)) +
geom_point() +
geom_errorbarh(aes(xmin = lower_CI, xmax = upper_CI), height = 0.2) +
geom_vline(xintercept = 1, linetype = "dashed") +
xlab("Odds Ratio (Treatment Effect)") +
ylab("Group") +
theme_minimal()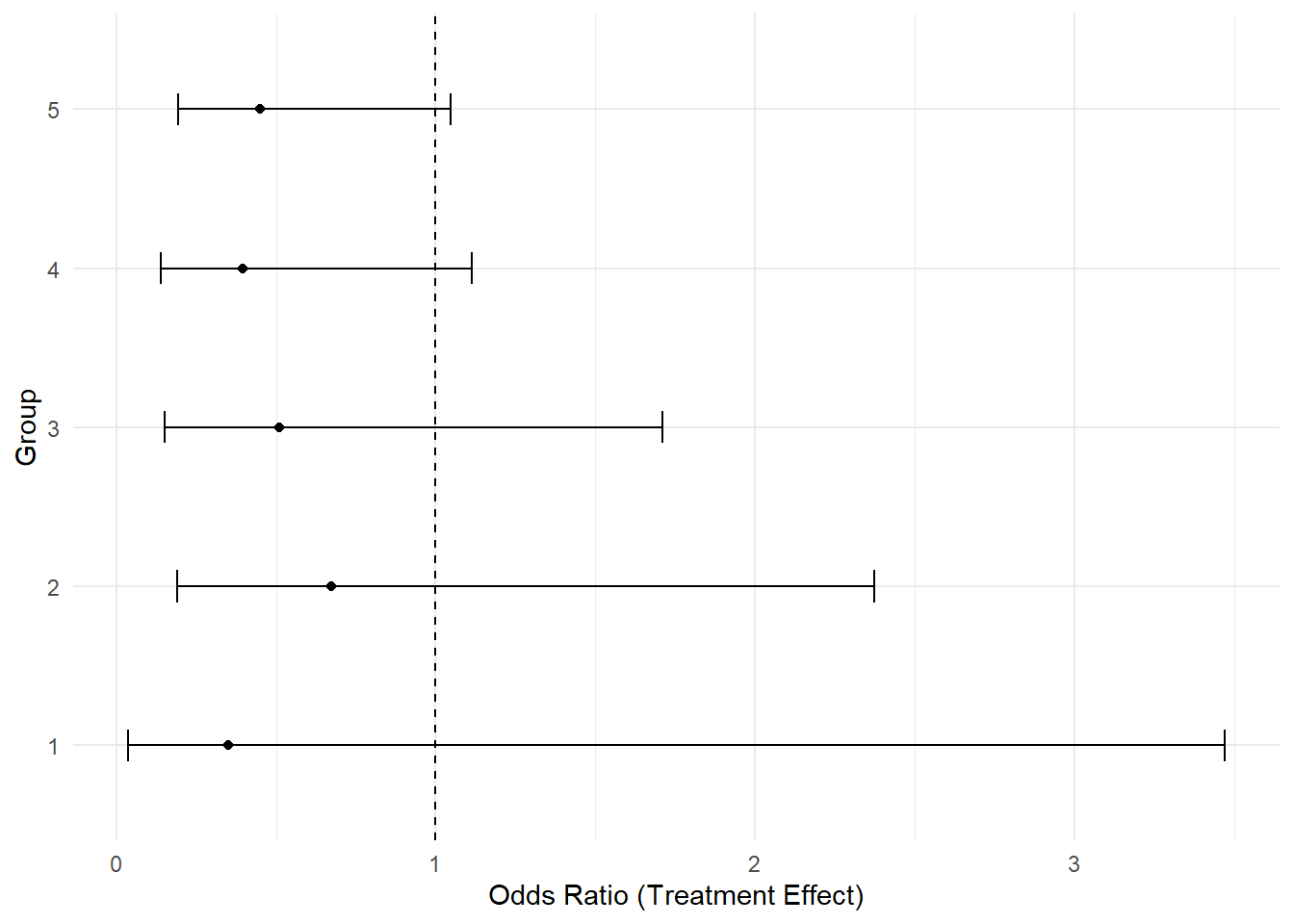
2. Hands-on excerise to run a causal forest
2.1 Prognostic Model (Random Forest)
library(randomForest)
rf_model <- randomForest(outcome ~ ., data = indo[, c(-1, -2)])
indo$rf_prob <- predict(rf_model, type = "response")2.2 Fit Causal Forest Model
# library(devtools)
# install_github("susanathey/causalTree")
library(causalTree)
cf_model <- causalForest(
formula = outcome ~ rx + rf_prob + age + risk + gender + sod + pep + recpanc + psphinc + precut + difcan + paninj + asa + sodsom + bsphinc + bstent + chole + train,
data = indo,
treatment = indo$rx,
ncov_sample = 10,
ncolx = 18,
split.Rule = "CT",
cv.option = "CT",
split.Honest = TRUE,
cv.Honest = TRUE,
split.Bucket = FALSE,
bucketNum = 5,
bucketMax = 100,
minsize = 20
)
indo$cARD <- predict(cf_model, newdata=indo)# Rank patients by treatment effect and group into quintile;
indo$cARDgp <- cut(
indo$cARD,
breaks = quantile(indo$cARD, probs = seq(0, 1, by = 0.2)),
include.lowest = TRUE,
labels = 1:5
)
# calculate mean and IQR of CARD for each group and plot it.
cARD_summary <- indo %>%
group_by(cARDgp) %>%
summarize(
median_cARD = median(cARD, na.rm = TRUE),
IQR_lower = quantile(cARD, 0.25, na.rm = TRUE),
IQR_upper = quantile(cARD, 0.75, na.rm = TRUE),
.groups = "drop"
)
print(cARD_summary)# A tibble: 5 × 4
cARDgp median_cARD IQR_lower IQR_upper
<fct> <dbl> <dbl> <dbl>
1 1 -0.0877 -0.0909 -0.0855
2 2 -0.0811 -0.0829 -0.0800
3 3 -0.0772 -0.0780 -0.0762
4 4 -0.0730 -0.0741 -0.0717
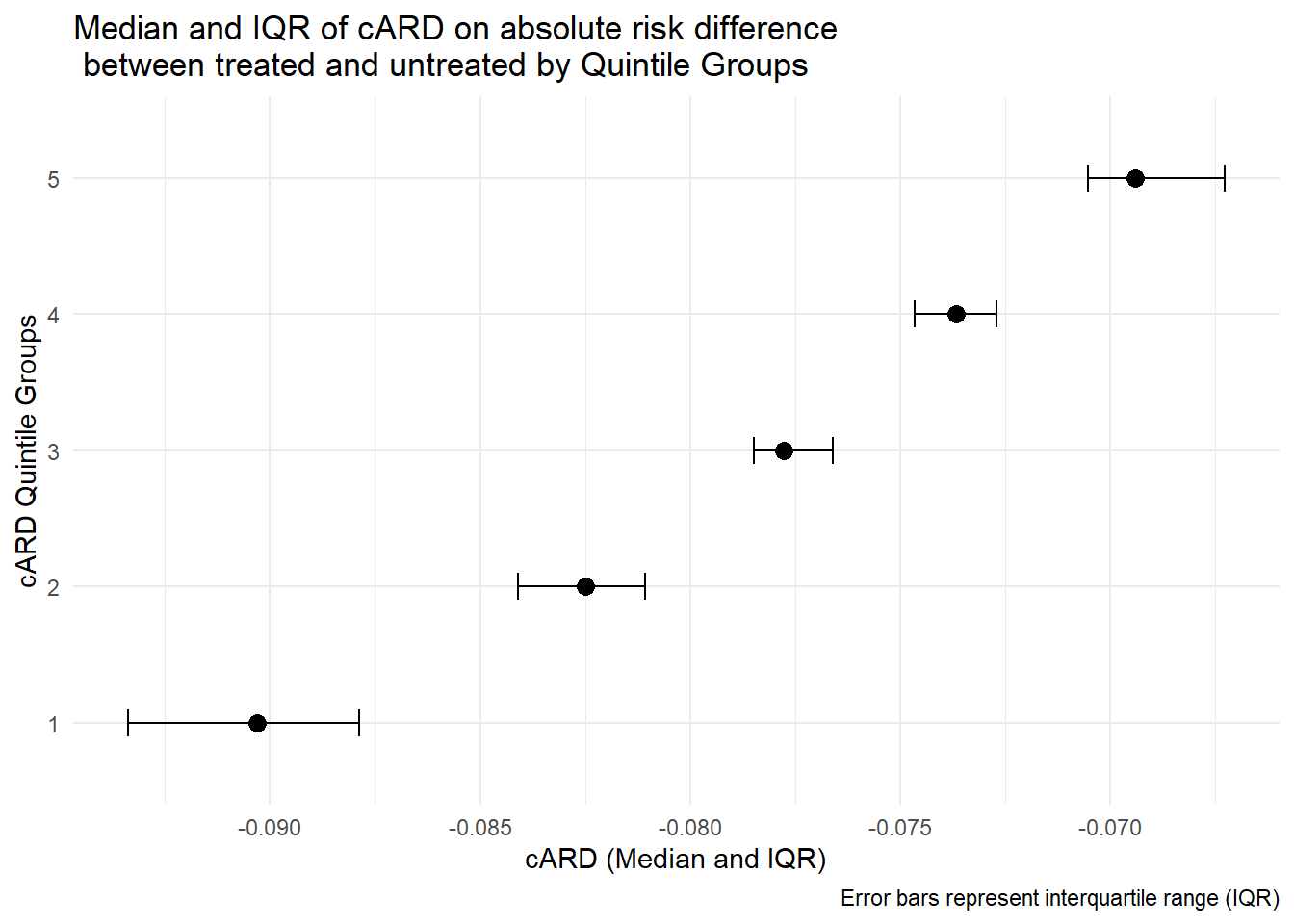
5 5 -0.0671 -0.0690 -0.0653# plot treatment effect by effect groups;
ggplot(cARD_summary, aes(y = cARDgp, x = median_cARD)) +
geom_point(size = 3) +
geom_errorbarh(aes(xmin = IQR_lower, xmax = IQR_upper), height = 0.2) +
theme_minimal() +
labs(
title = "Median and IQR of cARD on absolute risk difference \n between treated and untreated by Quintile Groups",
y = "cARD Quintile Groups",
x = "cARD (Median and IQR)",
caption = "Error bars represent interquartile range (IQR)"
)