library(tidyverse)
library(gtsummary)
library(DT)
options(scipen = 999)
hbv <- read.csv("HBV.csv", header = TRUE,fileEncoding="UTF-8-BOM")
# look at new data;
datatable(hbv,
rownames = FALSE,
options = list(dom = 't')) # display summary statistics by survival status;
hbv[,-1] %>%
tbl_summary(
by = HCC,
missing_text = "(Missing)") %>%
add_overall() %>%
add_p()| Characteristic | Overall N = 1611 |
0 N = 1081 |
1 N = 531 |
p-value2 |
|---|---|---|---|---|
| HBEAG | 45 (28%) | 31 (29%) | 14 (26%) | 0.8 |
| AGE | 47 (38, 56) | 46 (37, 54) | 53 (40, 60) | 0.005 |
| SEX | 129 (80%) | 89 (82%) | 40 (75%) | 0.3 |
| SPLENO | 46 (29%) | 23 (21%) | 23 (43%) | 0.004 |
| BILIRUB | 14 (10, 20) | 14 (10, 19) | 15 (10, 27) | 0.10 |
| (Missing) | 1 | 0 | 1 | |
| AST | 1.87 (1.00, 3.29) | 1.78 (1.00, 2.97) | 2.18 (1.43, 3.75) | 0.12 |
| ALT | 2.6 (1.2, 4.8) | 2.6 (1.5, 5.2) | 2.6 (1.0, 4.2) | 0.4 |
| PLAT | 146 (91, 195) | 158 (130, 203) | 95 (56, 140) | <0.001 |
| (Missing) | 1 | 1 | 0 | |
| ALBUMIN | 40 (36, 43) | 41 (38, 44) | 36 (32, 41) | <0.001 |
| (Missing) | 3 | 3 | 0 | |
| GAMMA | 17 (14, 22) | 15 (13, 21) | 19 (16, 25) | <0.001 |
| HCC_t | 72 (32, 98) | 82 (53, 106) | 40 (19, 72) | <0.001 |
| 1 n (%); Median (Q1, Q3) | ||||
| 2 Pearson’s Chi-squared test; Wilcoxon rank sum test | ||||
# checking out KM curve;
library(survival)
library(survminer)
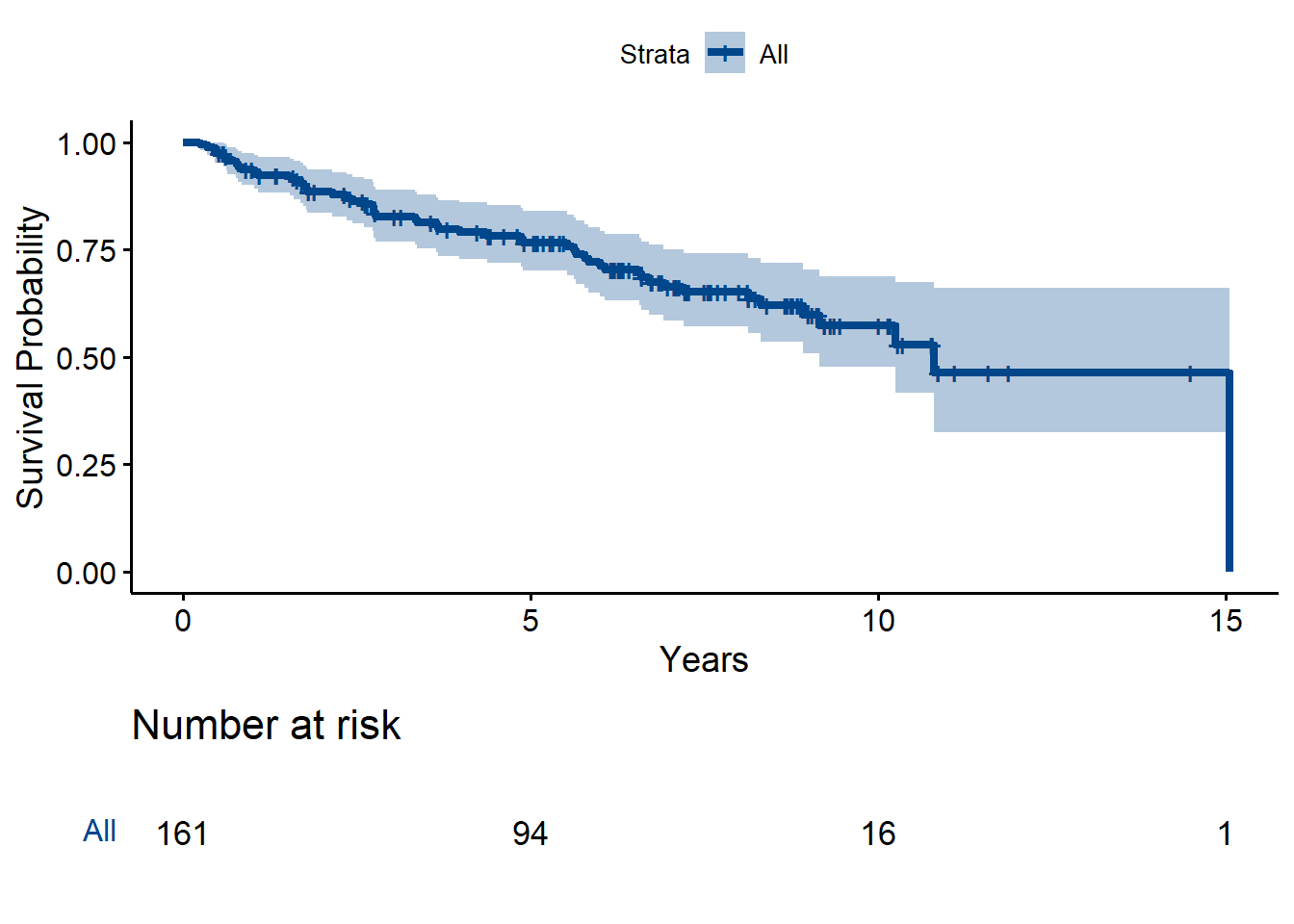
m0 <- survfit(Surv(HCC_t/12, HCC) ~ 1, data = hbv)
m0Call: survfit(formula = Surv(HCC_t/12, HCC) ~ 1, data = hbv)
n events median 0.95LCL 0.95UCL
[1,] 161 53 10.8 9.16 NAggsurvplot(m0,
data = hbv,
risk.table = TRUE,
size = 1.5,
palette = "lancet",
xlab = "Years",
ylab = "Survival Probability",
tables.theme = theme_cleantable())