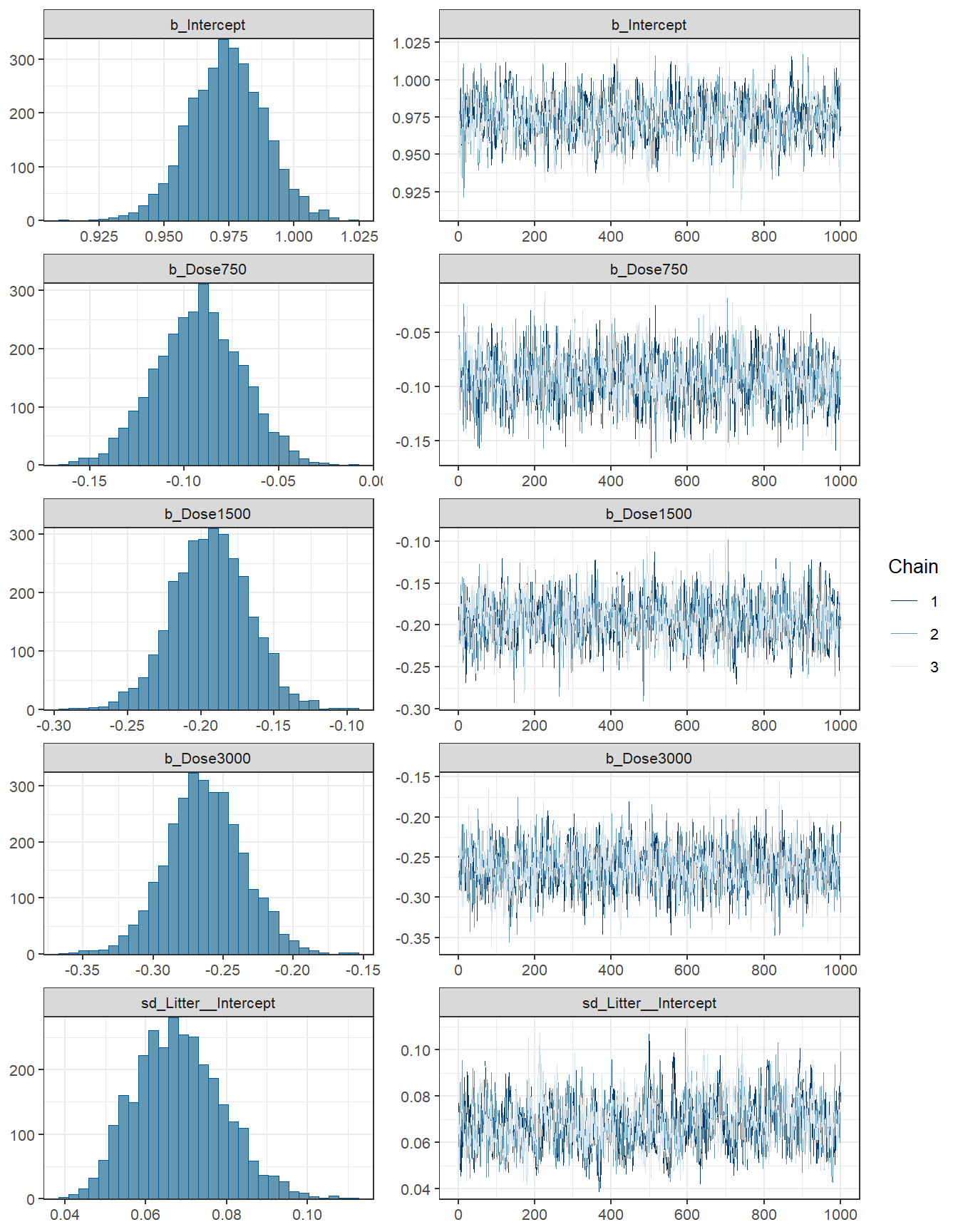
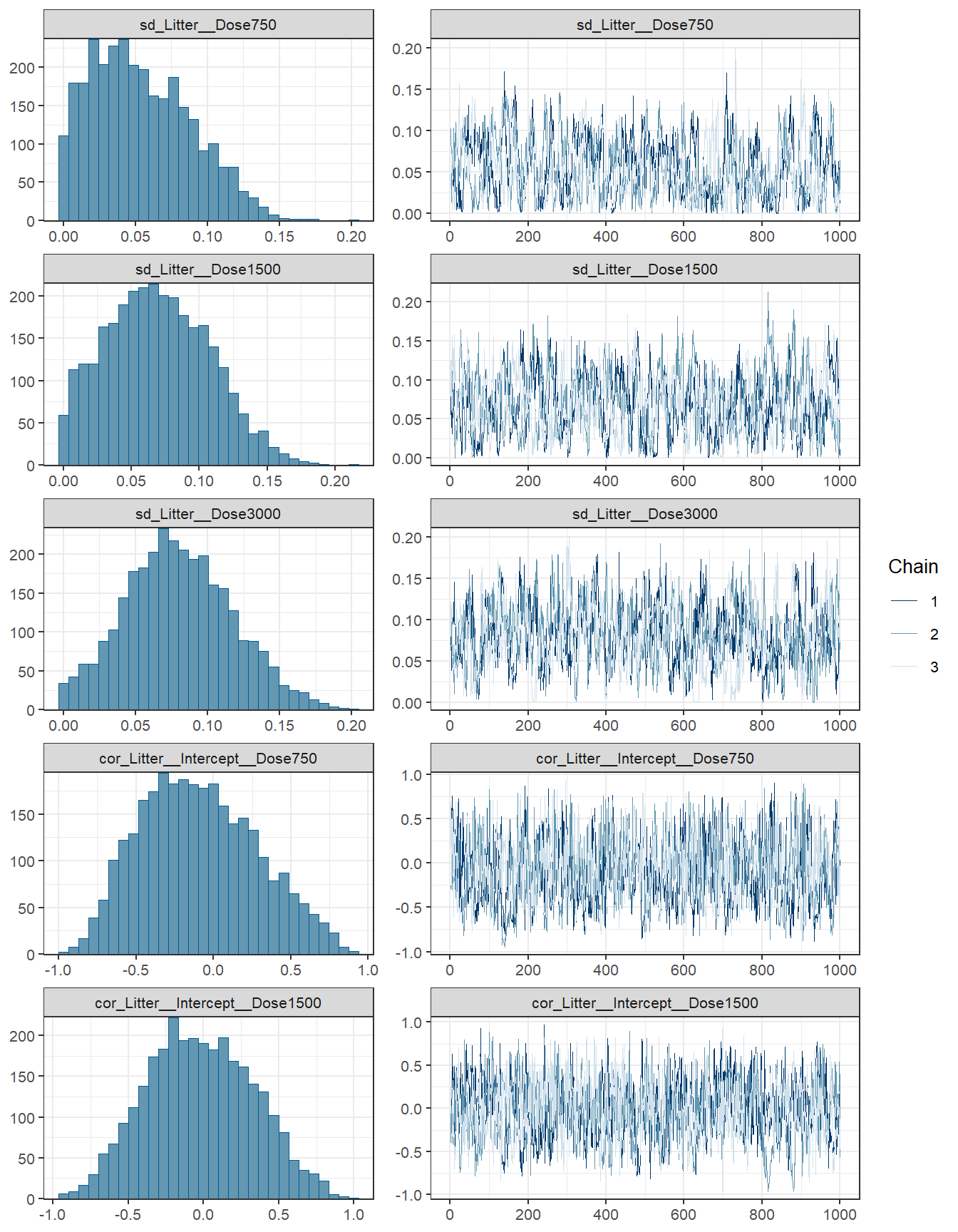
Bürkner, Paul-Christian. 2017. “Advanced Bayesian Multilevel Modeling with the r Package Brms.” arXiv Preprint arXiv:1705.11123.
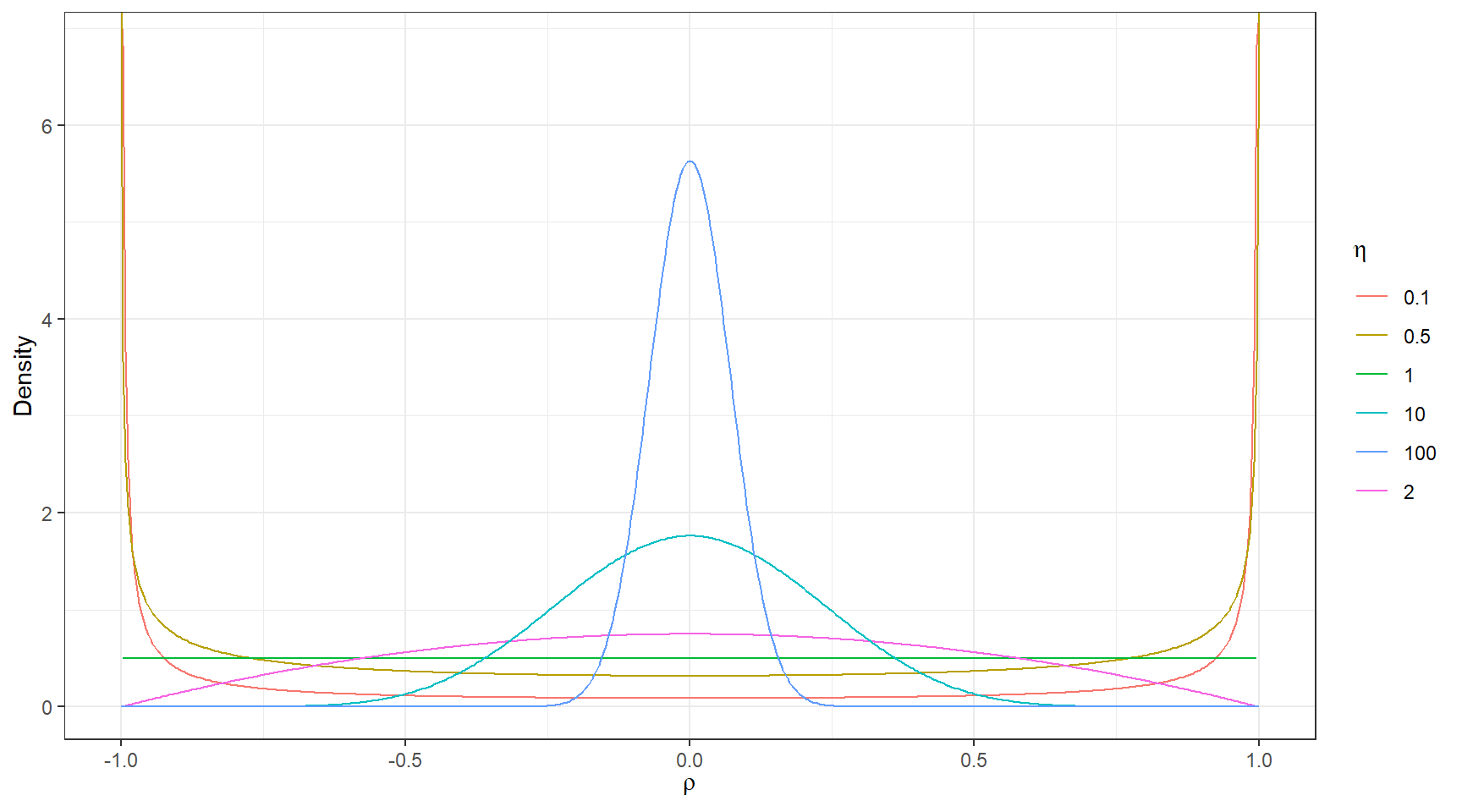
Lewandowski, Daniel, Dorota Kurowicka, and Harry Joe. 2009. “Generating Random Correlation Matrices Based on Vines and Extended Onion Method.” Journal of Multivariate Analysis 100 (9): 1989–2001.
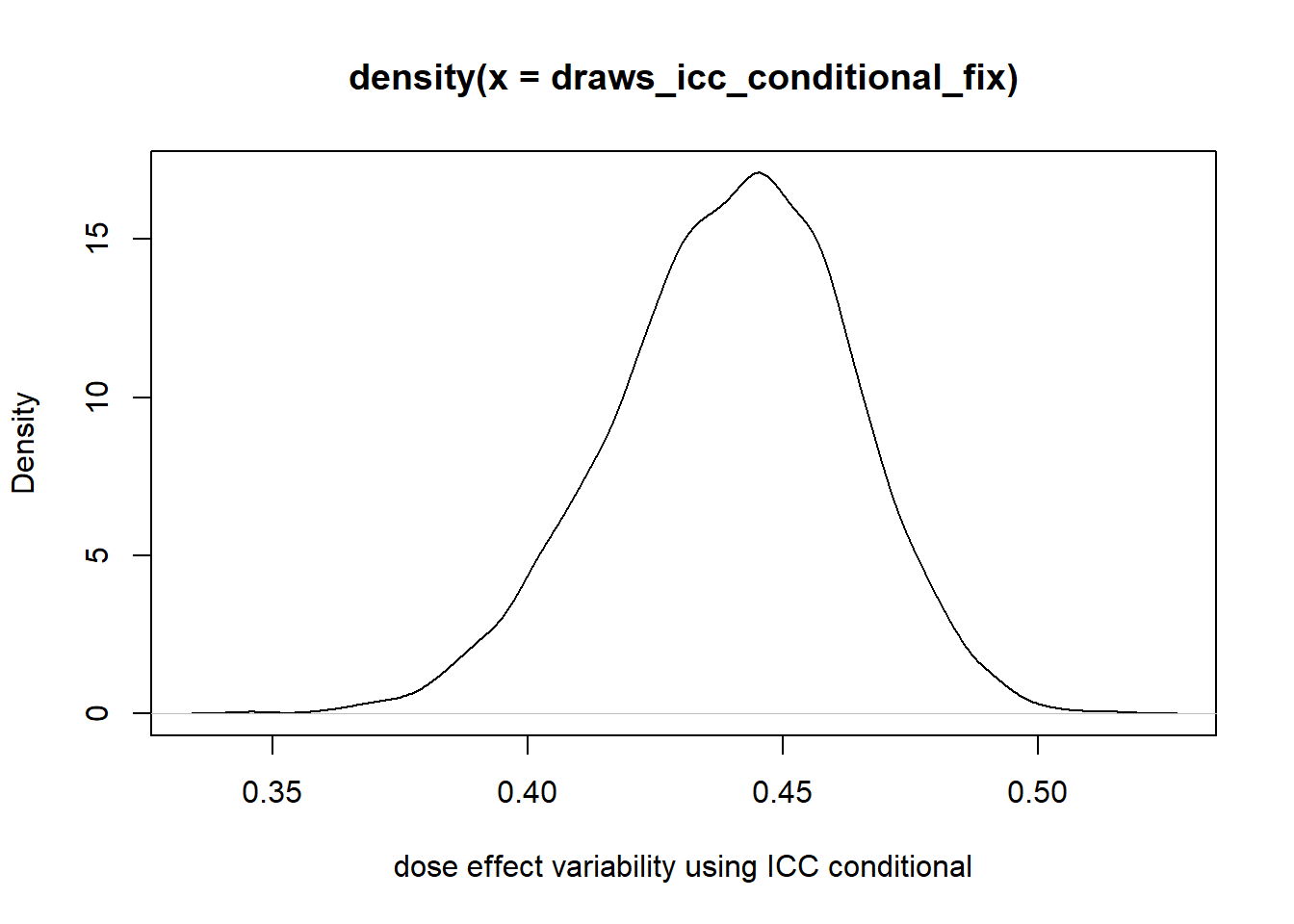
Nakagawa, Shinichi, Paul CD Johnson, and Holger Schielzeth. 2017. “The Coefficient of Determination r 2 and Intra-Class Correlation Coefficient from Generalized Linear Mixed-Effects Models Revisited and Expanded.” Journal of the Royal Society Interface 14 (134): 20170213.
Price, Catherine J, Carole A Kimmel, Rochelle W Tyl, and Melissa C Marr. 1985. “The Developmental Toxicity of Ethylene Glycol in Rats and Mice.” Toxicology and Applied Pharmacology 81 (1): 113–27.